Liquid biopsies have rapidly gained clinical interest, providing a minimally invasive means of acquiring and testing numerous biomarkers contained within blood and other bodily fluids. However, the analysis of circulating tumor (ct)DNA has unique considerations due to its low abundance. In oncology, its clinical implementation is hampered by the need for highly sensitive detection techniques. Addressing these challenges, droplet-based digital (dd)PCR enables absolute quantification of ctDNA with low detection limits. Even small sample quantities and rare genetic alterations may be reliably and precisely analyzed.
Does ctDNA testing have the potential to transform disease monitoring and treatment? To answer this, let’s first consider the limitations of conventional methods used in the clinical assessment of cancer. Then let us look at the capabilities of ddPCR solutions that are applicable to ctDNA analysis – and how recent technological advancements may support cancer research.
The diagnostic standard of care – and its limitations
Tissue biopsies and imaging protocols represent the current standard for cancer diagnosis, tumor profiling, and disease monitoring. However, these approaches have important limitations, including their inability to fully elucidate tumor heterogeneity, metastatic disease, molecular residual disease (MRD), or early molecular responses to therapy.
Traditional biopsies are invasive and can struggle to obtain tissue that is both of sufficient quantity and suitable quality. Further, a single biopsy sample may not be representative of the heterogeneity of the tumor as a whole or between multiple tumors, as in the case of metastatic disease. An analysis of urothelial carcinoma samples showed discordance in 23 percent of genomic alterations between primary and metastatic tumors derived from individual patients (1). Thus, performing repeated biopsies to evaluate metastasis and the treatment response over time may be impractical.
Imaging is most commonly used for diagnosis, tumor staging, and evaluation of disease progression. While easy to conduct and non-invasive, imaging insights rely on tumor size and growth rate. Techniques – such as X-ray, CT, MRI, ultrasound, and positron emission tomography – also vary in factors like resolution, sensitivity, and the need for contrast agents. They are differentially applied to meet specific goals of detecting, classifying, and monitoring disease.
However, achieving an optimal signal-to-noise ratio, reducing artifacts, and minimizing patient exposure to ionizing radiation from radioactive substances remain important considerations. In addition, solely tracking tumor dimensions may overlook changes occurring at the cellular and molecular level that provide key insights into the early response to treatment. Standard imaging techniques are also insufficiently sensitive for detecting MRD, which can predict disease recurrence.
ctDNA informs diagnosis, treatment response, and prognosis
Circulating cell-free DNA (cfDNA) is composed of double-stranded DNA fragments found outside of cells and has a varied half-life ranging from minutes to a few hours. cfDNA is released into the blood from numerous cell types, including immune cells, erythrocyte progenitors, hepatocytes, and vascular endothelial cells. The presence of cfDNA is the result of normal cellular processes, such as apoptosis, and its levels may rise during events that are associated with increased cell death, such as inflammatory responses.
ctDNA is a subset of cfDNA that is derived from tumor cells through apoptotic and necrotic processes, as well as cellular secretion. Useful clinical information may be obtained from ctDNA via the analysis of genetic mutations, methylation patterns, degree of fragmentation, and the overall ratio of ctDNA to cfDNA.
ctDNA may become an important cancer screening tool in the future, although this remains a challenge as it is present at very low levels in this setting. However, it may be used in certain scenarios, such as to complement standard diagnostic tests and confirm results, to identify at-risk individuals prior to performing invasive tests, or in cases that lack standardized screening methods.
In monitoring the response to treatment, MRD, and development of resistance to treatment, the utility of ctDNA is already evident. In fact, numerous assays have already been implemented in routine care (1). For example, some genomic profiling assays based on next-generation sequencing (NGS) are clinically validated and used to guide treatment decisions for solid tumors, such as non-small cell lung cancer (NSCLC) (2, 3).
The benefits of long-term ctDNA monitoring in NSCLC have also been demonstrated. Both NGS and ddPCR technologies have been used to evaluate the timing of the treatment response and the emergence of resistance mutations to tyrosine kinase inhibitors (4-7). Further, determining changes in ctDNA levels throughout the course of treatment relative to baseline may also serve as a prognostic marker across cancer types (8).
Improved detection limits resulting from advances in technologies such as NGS and ddPCR have bolstered the possibilities for ctDNA analysis in the context of managing cancer. NGS enables the simultaneous analysis of a multitude of epigenetic and genetic alterations, including unique mutations, and covers sequences ranging from kilobases to megabases.
However, NGS is time-intensive and may be cost prohibitive in situations where repeat analyses are required. Further, due to the breadth of data generated by NGS, their analysis and interpretation often requires support from biostatisticians. NGS is also associated with low-frequency errors, which are incurred during the PCR amplification step; thus, strategies such as the addition of molecular barcodes to DNA are often employed to differentiate real mutations from these errors.
ddPCR technology capabilities in ctDNA analysis
As an alternative to NGS, ddPCR technology provides more rapid results, and is both cost-effective and approachable in terms of workflow and data analysis (Figure 1). ddPCR technology is performed with low sample volumes and concentrations of nucleic acids, which are partitioned into thousands of nanoliter-sized droplets composed of a water-in-oil emulsion. This allows for absolute quantification of nucleic acids based on Poisson statistics and eliminates the reliance on a standard curve.
Low detection limits make ddPCR solutions suitable for various analyses, including rare genomic alterations, copy number variation, and DNA methylation patterns. Additionally, the low-cost and straightforward methodology of ddPCR technology warrants its use for routine disease monitoring.
Recent advancements in the field include capabilities such as seven color channels that enable researchers to design multiplex assays for simultaneous detection of numerous mutations. Other system features include automation as well as the requirement for low-volume sample input.
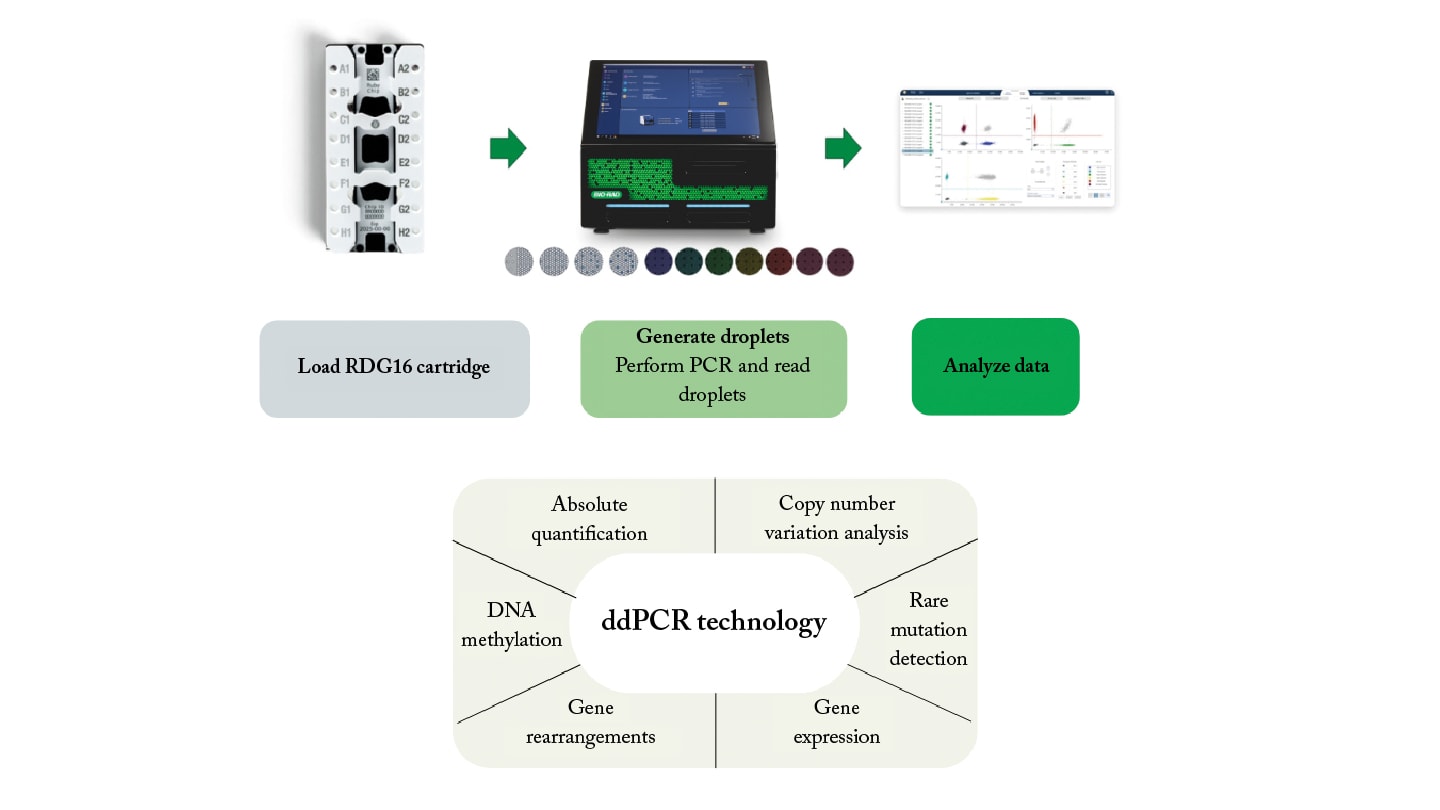
ddPCR technology in hematologic malignancies and solid tumor monitoring
Recently, ddPCR solutions have been employed for MRD analysis in lymphoproliferative disorders and the interrogation of characteristic alterations in follicular lymphoma (FL), chronic lymphocytic leukemia, mantle cell lymphoma (MCL), and Waldenstrom macroglobulinemia (10).
A comparative study of ddPCR technology versus real-time quantitative PCR (qPCR) was carried out in patients with FL, MCL, or multiple myeloma. MRD assessment in 26 samples deemed positive not-quantifiable by qPCR showed that 27 percent were quantifiable and 23 percent were negative by ddPCR (11).
A prospective, multicenter study evaluated the diagnostic potential of ctDNA mutation analysis (including EGFR, KRAS, and BRAF) by ddPCR for patients with primary lung cancer (12). In 142 patients with NSCLC and NGS results available from tissue biopsy, ddPCR assays detected 32 of 45 (71 percent) of driver mutations identified by NGS.
In two of those 142 patients, mutations were detected by ddPCR technology alone. Repeat biopsy followed by NGS confirmed the presence of an EGFR Ex19Del mutation detected solely by ddPCR technology, highlighting the potential of ddPCR solutions in capturing tumor heterogeneity.
The greater number of mutations detected by NGS overall was due to the ddPCR panel being limited to common and targetable mutations associated with lung cancer, which presents an opportunity for panel expansion.
In another recent study, multiplex ddPCR assays were evaluated in the detection of methylation markers across eight tumor types (including breast, colorectal, esophageal, head and neck, liver, lung, pancreatic, and prostate) (13).
Optimized duplex and triplex assays showed a significant difference in three methylation markers in tumor versus control tissue samples, except for one target in colorectal tissue. Combining targets increased the sensitivity and specificity of the assay, reaching a total cross-validated area under the curve of 0.948. This work paves the way for conducting such assays with liquid biopsies.
Conclusions
The treatment of cancer is increasingly moving towards a personalized approach, requiring advanced methods to support diagnosis, management of treatment, and disease monitoring over time. Traditional methods like tissue biopsies and imaging have limitations, such as invasiveness and variable sampling, and they may not reliably reflect tumor heterogeneity or the early changes associated with a therapeutic response.
ctDNA analysis in liquid biopsies is a minimally invasive and promising alternative, albeit one that requires highly sensitive detection techniques due to the low abundance of ctDNA. ddPCR technology offers a solution for ctDNA analysis, combining sensitivity for detection of rare genetic and epigenetic markers, multiplexing capabilities, cost-effectiveness, and a straightforward workflow. With established applications in oncology research, ddPCR technology is ripe with potential for adoption into clinical practice.
References
- A Bartolomucci et al., “Circulating tumor DNA to monitor treatment response in solid tumors and advance precision oncology,” NPJ Precis Oncol, 9, 1 (2025). PMID: 40122951
- JM Bauml et al., “Clinical validation of Guardant360 CDx as a blood-based companion diagnostic for sotorasib,” Lung Cancer, 166 (2022). PMID: 34838325.
- R Woodhouse et al., “Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin,” PLoS One, 15, 9 (2020). PMID: 32976510.
- ML Cheng et al., “Plasma ctDNA response is an early marker of treatment effect in advanced NSCLC,” JCO Precis Oncol, 5 (2021). PMID: 34250387
- I Dagogo-Jack et al., “Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA,” JCO Precis Oncol, 2018 (2018). PMID: 29376144.
- E Iwama et al., “Monitoring of somatic mutations in circulating cell-free DNA by digital PCR and next-generation sequencing during afatinib treatment in patients with lung adenocarcinoma positive for EGFR activating mutations,” Ann Oncol, 28, 1 (2017). PMID: 28177428.
- D Zheng et al., “Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance,” Sci Rep, 6 (2016). PMID: 26867973.
- DW Cescon et al., “Circulating tumor DNA and liquid biopsy in oncology,” Nat Cancer, 1, 3 (2020). PMID: 35122035.
- M Huerta et al., “Circulating tumor DNA detection by digital-droplet PCR in pancreatic ductal adenocarcinoma: a systematic review,” Cancers (Basel), 13, 5 (2021). PMID: 33673558.
- GM Assanto et al., “Research topic: measurable residual disease in hematologic malignancies. Can digital droplet PCR improve measurable residual disease monitoring in chronic lymphoid malignancies?” Front Oncol, 13 (2023). PMID: 36998457.
- D Drandi et al., “Minimal residual disease detection by droplet digital PCR in multiple myeloma, mantle cell lymphoma, and follicular lymphoma: a comparison with real-time PCR,” J Mol Diagn, 17, 6 (2015). PMID: 26319783.
- E Visser et al., “Up-front mutation detection in circulating tumor DNA by droplet digital PCR has added diagnostic value in lung cancer,” Transl Oncol, 27 (2023). PMID: 36413862.
- I Neefs et al., “Simultaneous detection of eight cancer types using a multiplex droplet digital PCR assay,” Mol Oncol, 19, 1 (2025). PMID: 39239847.




